Learn |

You’ve learned how to use pipes, and I’ve hyped a lot about how powerful they are. In this lesson, let’s see what my hype is all about. Let’s get you using pipes to do some complex things in fewer keystrokes and less time than you’d do with a GUI (if you could even do it in a GUI in the first place).
What you’ll learn
- The uniq command for grouping
- The sort command for sorting
- My tolerance for mixed metaphors
- Deeper composability
Computer scientists looooove a composable system. The command line is a composable system, it’s designed to leverage a little bit to get big work done. You’ve already dipped your toes in this composability. Composability on the command line is highly interactive, it encourages exploration to solve a problem. It’s easy to try things out quickly to see what works and what doesn’t, all with little consequence.
In this lesson, we’re getting shoulder-deep with composability. C’mon in, the water is refreshing.
Step 0: Before you start
Well, technically you’ve already started this lesson. But you should do and know a few things before you continue.
Do this: More cat, less trouble.
Step 1: Get this lesson’s files
You’ve been asked to make a comprehensive list of diseases that affect tomatoes. You’ve been given a set of CSV files, one file per type of disease. You need to combine these CSV files into a single CSV file.
Let’s get the files you’ll be working with.
Do this: In the shell, enter wget -qO- https://egopontem.com/lessons/sorting-grouping.tgz | tar -xz.
techwriter:~$ wget -qO- https://egopontem.com/lessons/sorting-grouping.tgz | tar -xz
techwriter:~$ ▮
Do this: Enter cd sorting-grouping.
techwriter:~$ cd sorting-grouping
techwriter:~/sorting-grouping$ ▮
Do this: Enter ls.
techwriter:~/sorting-grouping$ ls
README.txt bacteria.csv fungus.csv insect.csv
nematode.csv production.csv virus.csv
techwriter:~/sorting-grouping$ ▮
What happened: You downloaded this lesson’s files, switched to its directory, and listed the files in it.
Step 2: Tomatoes and oranges
English-speakers have an expression, “You can’t compare apples to oranges”. In our case, we have a bunch of CSV files. It doesn’t make sense to merge them unless they have the same columns. The first line of a CSV file lists its columns. Let’s look at the first lines of these CSV files to make sure they’re the same.
From what we learned in the previous lesson, we could use the less command to take a look at the first few lines of each CSV file. To check each CSV file, 6 in all, that’s a lot of typing.
What would be great is a command that shows just the first few lines of a file. And that command is head. Isn’t that great?
Let’s try it out with a single file to get a feel for it. By default, head outputs the first 10 lines of its input. There are a lot of fungi that attack tomatoes. What are the first 10?
Do this: Enter head fungus.csv
techwriter:~/sorting-grouping$ head fungus.csv
common,scientific
stem canker,Alternaria alternata
anthracnose,Colletotrichum
black mold rot,Stemphylium botryosum
black root rot,Thielaviopsis basicola
buckeye rot,Phytophthora
cercospora leaf mold,Pseudocercospora fuligena
charcoal rot,Macrophomina phaseolina
corky root rot,Pyrenochaeta lycopersici
didymella stem rot,Didymella lycopersici
techwriter:~/sorting-grouping$ ▮
Now let’s see the first 10 lines of all CSV files.
Do this: Enter head *.csv | less.
What happened: The head command shows a header for each file then the first 10 lines of that file. For example, one of headers from head looks like this: ==> bacteria.csv <==. And we piped this output to less to make it easier to scroll through the output.
Do this: Press q to return to the shell prompt when you’re done.
That was helpful, but we can get a more concise output that’s easier to read. We can also specify how many lines we want head to show with the -n option.
Do this: Enter head -n 1 *.csv | less.
What happened: We told head to show only the first line of each CSV file.
As we can see, our CSV files have the same columns except for production.csv, which has a column header that starts with country.
Do this: Press q to return to the shell prompt when you’re done.
There’s nothing wrong with a CSV file for tomato production. But we don’t want this information to screw up our merged diseases. Let’s just delete production.csv so that it stays out of our way.
Do this: Enter rm production.csv.
techwriter:~/sorting-grouping$ rm production.csv
techwriter:~/sorting-grouping$ ▮
What happened: We deleted production.csv. Let’s confirm that it’s not going to confuse our disease merger by repeating the previous command that shows the column headers for all CSV files.
Do this: Enter head -n 1 *.csv | less.
What happened: There. All tomatoes, no orange.
Do this: Press q to return to the shell prompt when you’re done.
We can go a step further by removing the file name header that head outputs. This is useful when we want to pipe the output from head to other commands that don’t need the headers. Really, head outputs the headers for people, not other commands. To to show you what that looks like, let’s try it out.
Do this: Enter head -q -n 1 *.csv.
techwriter:~/sorting-grouping$ head -q -n 1 *.csv
common,scientific
common,scientific
common,scientific
common,scientific
common,scientific
techwriter:~/sorting-grouping$ ▮
What happened: We’ve added an option to head. The -q option tells head not to output its file name header for each file. And notice that we’ve removed the pipe to less. We could have used the less command here, but we know that we’re dealing with just a handful of files, so we avoided the extra typing.
Step 3: Composability up to your ankles
But what if we really did have lots of CSV files with many columns? Double-checking a long list of column headers would get tedious. And the command line is all about removing the tedium.
What would be great is if there was a command that would single out the oranges among our tomatoes.
Do this: Enter head -q -n 1 *.csv | uniq.
techwriter:~/sorting-grouping$ head -n 1 *.csv | uniq
common,scientific
techwriter:~/sorting-grouping$ ▮
What happened: Look at that pipe, which is followed by uniq. We’ve composed some commands that show all the unique header lines in a set of CSV files. The uniq command shows only, well, the unique lines in its input. Since all the header lines are the same, we got just one line, which confirms that our CSV files have the same columns.
There’s a closely-related command that we normally use with uniq that we’ll get into later. For now, let’s just wade in a little deeper.
Step 3: Up to your knees
Now let’s merge the CSV files. If your first reaction is to use cat you’re on the right track. A command like cat *.csv does indeed just output all CSV files together.
But that’s not quite what we want. What we want, what we really, really want: Output a column header then output all tomato diseases. The first few lines of our merged CSV file should look something like this:
common,scientific
root-knot,Meloidogyne
sting,Belonolaimus longicaudatus
stubby-root,Paratrichodorus
Our challenge is that each CSV file has its own column header, which the cat command would output.
Do this: Enter cat *.csv | less.
What happened: See what I mean? The column header shows up every few lines. We only want it to appear once at the beginning of our merged output. And lucky us, we already know how to output the column header by itself.
Do this: Press q to return to the shell prompt when you’re done.
Now we need to output the contents of our CSV files without their column headers. We’ll use tail for that. The tail command does what you’re probably guessing: it outputs the last few lines of a file.
Just like head, tail outputs the last 10 lines of a file by default. And we can override that default with the -n option.
Do this: Enter tail -q -n 1 *.csv.
techwriter:~/sorting-grouping$ tail -q -n 1 *.csv
tomato big bud,Phytoplasma
white mold,Sclerotinia sclerotiorum
tomato pinworm,Keiferia lycopersicella
stubby-root,Paratrichodorus
tomato planto macho,Tomato planto macho viroid
techwriter:~/sorting-grouping$ ▮
What happened: We asked tail to output the last line of each CSV file with no file name headers. This isn’t it exactly what we want, but now you know that tail works the same way as head, only from the other end of its input.
Now let’s tell tail to output all but the first line of each CSV file. To do this, we still use the -n option, but we specify its numerical value a little differently. Just to try this out, we’ll use a single, short CSV file.
First let’s see all of nematode.csv.
Do this: cat nematode.csv.
techwriter:~/sorting-grouping$ cat nematode.csv
common,scientific
root-knot,Meloidogyne
sting,Belonolaimus longicaudatus
stubby-root,Paratrichodorus
techwriter:~/sorting-grouping$ ▮
What happened: There’s all of the contents of nematode.csv. Pay attention to the first line, which is the list of columns, common,scientific.
Now let’s see the contents of nematode.csv starting from its 2nd line.
Do this: tail -q -n +2 nematode.csv.
techwriter:~/sorting-grouping$ tail -q -n +2 nematode.csv
root-knot,Meloidogyne
sting,Belonolaimus longicaudatus
stubby-root,Paratrichodorus
techwriter:~/sorting-grouping$ ▮
What happened: The -n +2 option means “Show a file’s contents starting from line 2 all the way to the end”. In other words, we told tail to show all of a CSV file’s contents except for its column header.
Step 4: Up to your elbows
Ok! Now we know how to show just the column header for a bunch of similarly-columned CSV files:
head -q -n 1 *.csv | uniq
And we know how to show only the data rows for some CSV files:
tail -q -n+2 *.csv
Let’s compose these two things. Yup, compose. In practice, composition on the command line just means stringing together small changes into a bigger change.
One way we learned to compose is to use pipes. A pipe connects output from one command directly to the input of the next command. But to compose the command for the column header and the command for the CSV rows, a pipe won’t do.
Try it out to find out.
Do this: Enter head -q -n 1 *.csv | uniq | tail -q -n+2 *.csv | less.
What happened: The less command shows the first screenful. Notice that the first line isn’t the column header, which means the output that less shows doesn’t include head -q -n 1 *.csv | uniq.
And that’s not what we want. And that’s because we’ve told the shell to take the output from head, feed it into uniq, then take that output to feed it into tail. But we also told tail to read from CSV files instead of standard input. And that’s what tail did, completely ignoring the output from uniq, which isn’t what we would have wanted tail to process anyway. So less ends up only showing what tail outputs.

What we want is a way to tell the shell to do this:

- Generate output for the column headers with head and uniq.
- And then generate output for the data rows with tail.
Not only do we want to keep the output from the first command, we don’t want this output to interfere with the input of the second command. But we want the output from both commands to be treated as if it were coming from a single command so that we can further process its output for commands like less. (Whew! Read that again if you have to.)
The way we’ll do that is with &&. The && is an operator in the shell that we use to combine commands in exactly the way I just described.
To keep things simple to see how && works, let’s try it with just a single CSV file.
Do this: Enter head -q -n 1 nematode.csv | uniq && tail -q -n+2 nematode.csv.
techwriter:~/sorting-grouping$ head -q -n 1 nematode.csv | uniq && tail -q -n+2 nematode.csv
common,scientific
root-knot,Meloidogyne
sting,Belonolaimus longicaudatus
stubby-root,Paratrichodorus
techwriter:~/sorting-grouping$ ▮
What happened: Isn’t that something? We see the column header, and then the data rows, in nematode.csv.
The && operator tells the shell to run the first command, in our case this is head -q -n 1 nematode.csv | uniq. Then, if this first command runs without errors, the && operator tells the shell to run the next command, tail -q -n+2 nematode.csv. The important thing to take away from && is that it doesn’t connect command output and input in any way, it just runs one command and then the next.
The effect is that we get the output of the first command and then the output of the next command, and neither command has an effect on the other’s standard intput or output. And that’s exactly what we want.
Yes, we’re using | uniq redundantly here. I’ll let you figure out why yourself. But the advantage here is that this confirms that we can get the column header with one file or many.
Now let’s apply this to all CSV files. This next command is a long one, so copy and paste it carefully.
Do this: Enter ( head -q -n 1 *.csv | uniq && tail -q -n+2 *.csv ) | less
What happened: The less command is the last of this composition. It’s showing us a single column header, common,scientific, followed by a bunch of tomato diseases. Success!
Do this: Press q to return to the shell prompt when you’re done.
Let’s unpack this command:
- The opening and closing parentheses, ( and ), mean “Treat whatever happens inside these parentheses as if it were a single command”.
- Inside the parentheses we use the && to generate the column header output and then the data row output. We are intentionally not connecting the output from one to the other.
- After the ), the | less just pipes to less. So effectively, less gets output from whatever is generated inside the parentheses.
There’s an extra detail that you should know about the parentheses. What they really mean is to tell the shell to run the commands within the parentheses in a sub-shell.
Ask me what a sub-shell is. I’ll answer you even if you didn’t ask: a sub-shell is another shell that the shell runs. Yes, the shell can run another shell just like it can run other commands. It can do this because the shell is a command itself, with its own standard input and standard output that can be redirected.
The reason I bring up this detail is that I hope it clarifies what the ( and ) do. The reason we can pipe to less with the output from the commands in ( and ) is that we’re actually getting output from a sub-shell. And the | less is actually piping the sub-shell’s output to the input for less.
Conceptually, this is what we’ve done:
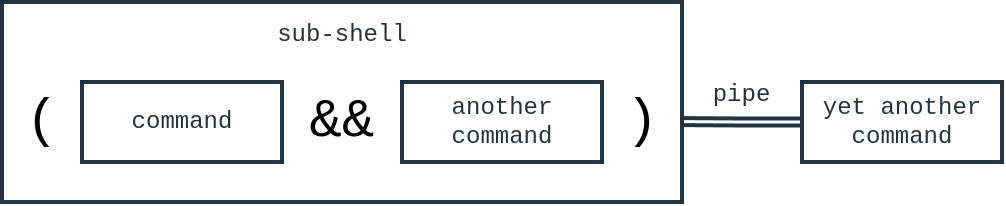
Whatever is in the ( and ) is up to you. You can even put a single command, like just cat for example. Really, the most common use of the sub-shell is to treat a bunch of commands as if they were a single command so that it’s easier to redirect input and output to a single place.
Let that sit in your head a bit before you continue.
Step 5: Now the shoulders
Now we have a well-formed CSV file:
- The first line is the column header.
- The rest of the file lists the data rows for our tomato diseases.
Nice work! Except for one thing. Just a small thing. Wouldn’t it be nice if the list of tomato diseases was in alphabetical order? Of course it would. If you’re surprised that there’s a command for sorting, then I’m surprised because it shouldn’t be surprising.
That command is sort. It takes its input, rearranges its lines alphabetically, then outputs the result. Let’s keep building on what we’ve already done. Composability for the win! The question now is to figure out where to put sort. We want to sort the data rows of our merged CSV file without the column header. That means we want to sort the output of the tail command.
Do this: Enter ( head -q -n 1 *.csv | uniq && tail -q -n+2 *.csv | sort ) | less
What happened: The less command shows the column header followed by data rows sorted alphabetically. Another success!
Do this: Press q to return to the shell prompt when you’re done.
Step 6: Don’t uniq until you sort
The uniq command is great for grouping, too. You can use it to show what groups are in your input. But that depends on using it properly. There’s something important about uniq that you should know. The uniq command shows unique lines when they are adjacent. In other words, it doesn’t detect unique lines if they aren’t sorted first. In our case, earlier we got away with using this command despite not sorting input: head -q -n 1 *.csv | uniq.
In the spirit of no-consequence, interactive exploration, we just wanted to get a quick confirmation that our column headers are the same. Any output of more than one line quickly let us know that the columns headers in the CSV files were not the same.
But to use uniq reliably, it’s a best practice to first sort its input. Get in the habit of using | sort | uniq instead of just | uniq.
Do this: Enter head -q -n 1 *.csv | sort | uniq.
techwriter:~/sorting-grouping$ head -q -n 1 *.csv | sort | uniq
common,scientific
techwriter:~/sorting-grouping$ ▮
What happened: We added a piped command, sort. We’ve added this pipe before uniq to let it find unique adjacent lines.
Of course, the result for us, this time, is the same. But just keep this in mind when using uniq: Make sure its input is sorted first.
You did it!
Congratulations, you know how to compose basic commands into some pretty sophisticated processing.
What you learned
- tail is the counterpart to head. We can use tail to show the last lines of its input. And we can use it to ignore the beginning lines of input.
- By the way, the same goes for head. We can use it show the beginning lines of its input. And we can use head to ignore the last lines of input.
- uniq collapses identical, adjacent lines into a single line. It’s great for grouping identical lines of text. For accurate results, input from uniq should come from output from sort. In other words, if you’re going to use uniq, use sort | uniq instead.
- sort rearranges its lines of input alphabetically.
- | lets us string together commands by connecting one command’s output to the next command’s input.
- On the other hand, && lets us string together commands by simply running one then the next without tying their input or output together.
- Parentheses, ( and ) let us treat a bunch of commands as if they are a single command. It does this with a sub-shell. Because the sub-shell is itself just a command, we can redirect its input and output like any other command.